What data is to be fit?¶
The Sherpa Data class is used to
carry around the data to be fit: this includes the independent
axis (or axes), the dependent axis (the data), and any
necessary metadata. Although the design of Sherpa supports
multiple-dimensional data sets, the current classes only
support one- and two-dimensional data sets.
Overview¶
The following modules are assumed to have been imported:
>>> import numpy as np
>>> import matplotlib.pyplot as plt
>>> from sherpa.stats import LeastSq
>>> from sherpa.optmethods import LevMar
>>> from sherpa import data
Names¶
The first argument to any of the Data classes is the name
of the data set. This is used for display purposes only,
and can be useful to identify which data set is in use.
It is stored in the name attribute of the object, and
can be changed at any time.
The independent axis¶
The independent axis - or axes - of a data set define the
grid over which the model is to be evaluated. It is referred
to as x, x0, x1, ... depending on the dimensionality
of the data (for
binned datasets there are lo
and hi variants).
Although dense multi-dimensional data sets can be stored as arrays with dimensionality greater than one, the internal representation used by Sherpa is often a flattened - i.e. one-dimensional - version.
The dependent axis¶
This refers to the data being fit, and is referred to as y.
Unbinned data¶
Unbinned data sets - defined by classes which do not end in
the name Int - represent point values; that is, the the data
value is the value at the coordinates given by the independent
axis.
Examples of unbinned data classes are
Data1D and Data2D.
>>> np.random.seed(0)
>>> x = np.arange(20, 40, 0.5)
>>> y = x**2 + np.random.normal(0, 10, size=x.size)
>>> d1 = data.Data1D('test', x, y)
>>> print(d1)
name = test
x = Float64[40]
y = Float64[40]
staterror = None
syserror = None
Binned data¶
Binned data sets represent values that are defined over a range,
such as a histogram.
The integrated model classes end in Int: examples are
Data1DInt
and Data2DInt.
It can be a useful optimisation to treat a binned data set as an unbinned one, since it avoids having to estimate the integral of the model over each bin. It depends in part on how the bin size compares to the scale over which the model changes.
>>> z = np.random.gamma(20, scale=0.5, size=1000)
>>> (y, edges) = np.histogram(z)
>>> d2 = data.Data1DInt('gamma', edges[:-1], edges[1:], y)
>>> print(d2)
name = gamma
xlo = Float64[10]
xhi = Float64[10]
y = Int64[10]
staterror = None
syserror = None
>>> plt.bar(d2.xlo, d2.y, d2.xhi - d2.xlo);
Errors¶
There is support for both statistical and systematic
errors by either using the staterror and syserror
parameters when creating the data object, or by changing the
staterror and
syserror attributes of the object.
Filtering data¶
Sherpa supports filtering data sets; that is, temporarily removing parts of the data (perhaps because there are problems, or to help restrict parameter values).
The mask attribute indicates
whether a filter has been applied: if it returns True then
no filter is set, otherwise it is a bool array
where False values indicate those elements that are to be
ignored. The ignore() and
notice() methods are used to
define the ranges to exclude or include. For example, the following
hides those values where the independent axis values are between
21.2 and 22.8:
>>> d1.ignore(21.2, 22.8)
>>> d1.x[np.invert(d1.mask)]
array([ 21.5, 22. , 22.5])
After this, a fit to the data will ignore these values, as shown
below, where the number of degrees of freedom of the first fit,
which uses the filtered data, is three less than the fit to the
full data set (the call to
notice() removes the filter since
no arguments were given):
>>> from sherpa.models import Polynom1D
>>> from sherpa.fit import Fit
>>> mdl = Polynom1D()
>>> mdl.c2.thaw()
>>> fit = Fit(d1, mdl, stat=LeastSq(), method=LevMar())
>>> res1 = fit.fit()
>>> d1.notice()
>>> res2 = fit.fit()
>>> print("Degrees of freedom: {} vs {}".format(res1.dof, res2.dof))
Degrees of freedom: 35 vs 38
Note
It’s a bit confusing since not always clear where a given attribute or method is defined. There’s also get_filter/get_filter_expr to deal with. Also filter/apply_filter.
Visualizing a data set¶
The data objects contain several methods which can be used to
visualize the data, but do not provide any direct plotting
or display capabilities. There are low-level routines which
provide access to the data - these include the
to_plot() and
to_contour() methods - but the
preferred approach is to use the classes defined in the
sherpa.plot module, which are described in the
visualization section:
>>> from sherpa.plot import DataPlot
>>> pdata = DataPlot()
>>> pdata.prepare(d2)
>>> pdata.plot()
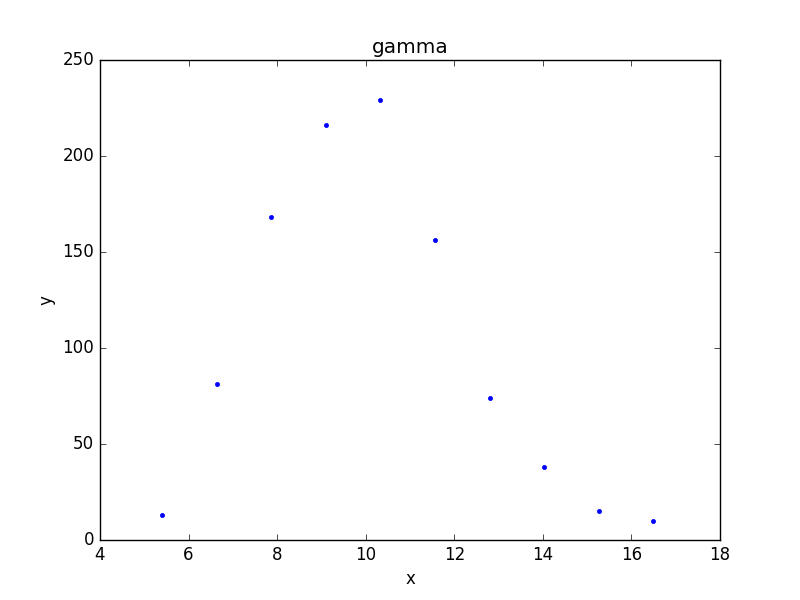
Although the data represented by d2 is
a histogram, the values are displayed at the center of the bin.
The plot objects automatically handle any filters applied to the data, as shown below.
>>> d1.ignore(25, 30)
>>> d1.notice(26, 27)
>>> pdata.prepare(d1)
>>> pdata.plot()
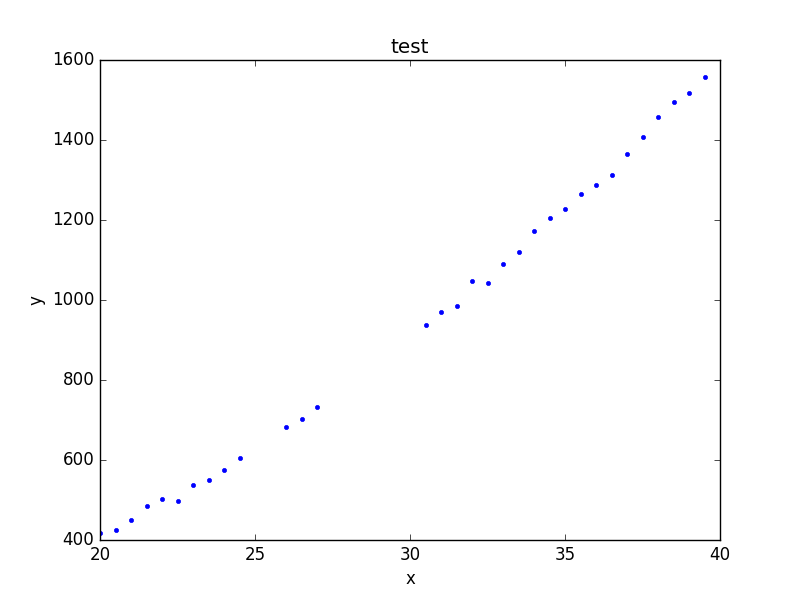
Note
The plot object stores the data given in the
prepare() call,
so that changes to the underlying objects will not be reflected
in future calls to
plot()
unless a new call to
prepare() is made.
>>> d1.notice()
At this point, a call to pdata.plot() would re-create the previous
plot, even though the filter has been removed from the underlying
data object.
Evaluating a model¶
The eval_model() and
eval_model_to_fit()
methods can be used
to evaluate a model on the grid defined by the data set. The
first version uses the full grid, whereas the second respects
any filtering applied to the data.
>>> d1.notice(22, 25)
>>> y1 = d1.eval_model(mdl)
>>> y2 = d1.eval_model_to_fit(mdl)
>>> x2 = d1.x[d1.mask]
>>> plt.plot(d1.x, d1.y, 'ko', label='Data')
>>> plt.plot(d1.x, y1, '--', label='Model (all points)')
>>> plt.plot(x2, y2, linewidth=2, label='Model (filtered)')
>>> plt.legend(loc=2)
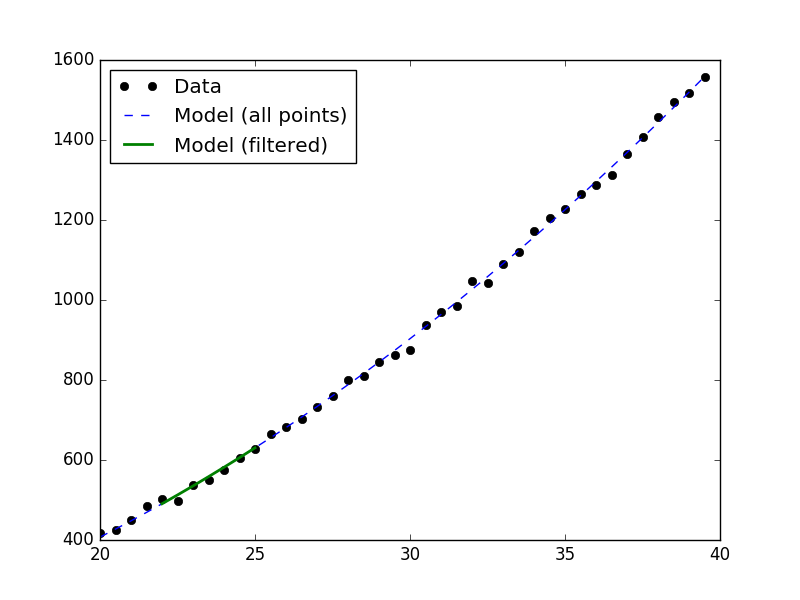
Reference/API¶
Note
There are a bunch of methods that seem out of place; e.g. Data1D has get_x0 even though it’s not ND!