Simulation for XMM spectra (AO-4)
Last modified:
February 26, 2007
This thread uses
PINTofALE
to generate simulated XMM EPIC-PN, EPIC-MOS, and RGS spectra
of a coronal source normalized to a ROSAT/PSPC count rate.
The XMM ARF, RMF and RSP files used here can be obtained from the
PROPOSAL & PLANNING TOOLS page.
- Initialize variables
- Define a DEM
- Rescale DEM to match ROSAT count rate
- Construct spectra
- Simulate and plot
- Comparison with PIMMS
All the steps described here are repeated in the .par file:
XMM_epic_rgs_ao4.par.
0. Initialize the settings
# start IDL
idl
; set up the PoA environment
;
.run initale
;NOTE:
; if INITALE fails, run the script
; @PoA_constructor
1. Initialize some variables
; Because some variables will be used repeatedly in the course of this
; thread, it might be useful to initialize them now. Nailing down these
; variables now will also allow for something of a checklist for some
; important quantities/files needed for the task at hand:
; Local Environment We will set up the pathnames specific to
; the local installation here
!LDBDIR = '$CHIANTI' ; Atomic Line Database
; choose from the predefined '$CHIANTI', '$SPEX', '$APED',
; or specify the full path name to the line database
pimmsdir='/soft/pimms/data' ; the full path name to the PIMMS database
; Source We will characterize our model source by the following:
!NH = 3e20 ; H column density [cm^-2]
!EDENS = 1.0e9 ; electron number density [cm^-3]
!ABUND = getabund('grevesse et al.') ; element abundances
;(SEE: getabund())
T_components = [6.1, 6.8, 7.2] ; log(T[K]) components in EM
EM_components = [6.1d11, 6.1d11, 7.1e11] ; Emission Measure [cm^-3]
; Observation The observation can be described by:
EXPTIME = 50. ; nominal exposure time [ks]
obs_rate = 0.1 ; ROSAT/PSPC count rate [ct/s]
; (set to 0 or less to be ignored)
pn_ARF = 'pn-medium.arf' ; EPIC-PN ARF filename or 'none'
mos_ARF = 'mos1-medium.arf' ; EPIC-MOS ARF filename or 'none'
rgs1_ARF = 'none' ; RGS1 ARF filename or 'none'
rgs2_ARF = 'none' ; RGS2 ARF filename or 'none'
pn_RMF = 'pn.rmf' ; EPIC-PN RMF filename
mos_RMF = 'mos1.rmf' ; EPIC-MOS RMF filename
rgs1_RMF = 'RGS1ORDER1.RSP' ; RGS-1 RMF filename
rgs2_RMF = 'RGS2ORDER1.RSP' ; RGS-2 RMF filename
;NOTE: the standard RGS response matrices include the
;effective area, and hence an ARF should not be specified for them.
; Analysis Parameters
; We may restrict the analysis to the wavelength ranges of interest.
; Here we set the ROSAT/PSPC passband used to measure obs_rate,
; the desired XMM output passband, and the wavelength grid
; for the idealized model spectrum. The output spectra will be
; defined on the default response energy grids restricted to
; the ranges defined here.
emin_rosat = 0.12 ; minimum energy in ROSAT/PSPC passband [keV]
emax_rosat = 2.48 ; maximum energy in ROSAT/PSPC passband [keV]
emin_xmm = 0.2 ; minimum energy in XMM passband [keV]
emax_xmm = 8.0 ; maximum energy in XMM passband [keV]
wmin_rosat = !fundae.kevang/emax_rosat ; convert to [Ang]
wmax_rosat = !fundae.kevang/emin_rosat ; convert to [Ang]
wmin_xmm = !fundae.kevang/emax_xmm ; convert to [Ang]
wmax_xmm = !fundae.kevang/emin_xmm ; convert to [Ang]
!WMIN = wmin_rosat < wmin_xmm ; minimum wavelength for model [Ang]
!WMAX = wmax_rosat > wmax_xmm ; maximum wavelength for model [Ang]
nwbin = 10000L ; number of bins in model spectrum
2. Define a DEM
; A Differential Emission Measure (DEM) is required to estimate the
; amount of emission at various temperatures. Typically, a 2-temperature
; model is used. Here we will use PINTofALE's mk_dem(), which constructs
; a DEM array given a temperature grid and emission measure components.
; We use as the temperature grid !LOGT. The emission measure components
; are T_components and EM_components as defined above. We choose 'delta'
; to treat the EM components as delta functions (see mk_dem() or the
; Chandra/ACIS example for more options)
!DEM=mk_dem('delta', logT = !LOGT, pardem=T_components, indem=EM_components)
3. Rescale the DEM to match ROSAT count rate
; We will assume that a ROSAT/PSPC count rate is available,
; and that the simulation will match this rate. We will assume
; a count rate of 0.1 cts/s . Data from other missions such
; as ASCA, BeppoSAX, etc. can be dealt with in the same manner.
; First find and read in the ROSAT/PSPC effective area.
; You will need to know where your local PIMMS installation
; is to do this.
rosat_pspc_open=get_pimms_file('ROSAT','PSPC','OPEN',pdir=pimmsdir)
rd_pimms_file, rosat_pspc_open, pspc_effar, pspc_wvlar, /wave
; Make sure that the wavelengths are sorted in increasing order
ae=sort(pspc_wvlar) & pspc_wvlar=pspc_wvlar[ae] & pspc_effar=pspc_effar[ae]
; The following is similar to the process described in the detailed
; example thread (see Section 1 and Section 2)
A] Read in line cooling emissivities and calculate line intensities
; Read line cooling emissivities of all possible lines in the
; ROSAT/PSPC wavelength range from the atomic data base.
; NOTE: To avoid multiple reads of the line emissivity database, we
; shall read in the emissivities over the entire range of interest
lconf=rd_line(atom,n_e=!EDENS,$
wrange=[MIN(pspc_wvlar)<!WMIN,MAX(pspc_wvlar)>!WMAX],$
dbdir=!LDBDIR,verbose=!VERBOSE,wvl=LWVL,logT=LLOGT,Z=Z,$
ion=ION,jon=JON,fstr=lstr)
; The output of rd_line() will only include level population,
; and not ion balances. We will use fold_ioneq() to fold ion balances.
; NOTE: This step should not be performed if !LDBDIR is set to APED,
; which already includes ion balances and abundances.
if strpos(strlowcase(!LDBDIR),'aped',0) lt 0 then lconf=$
fold_ioneq(lconf,Z,JON,chidir=!CHIDIR,$
logT=LLOGT,eqfile=!IONEQF,verbose=!VERBOSE)
; And now calculate line intensities using lineflx().
v_ABUND = !ABUND
; NOTE: If !LDBDIR is set to APED, Anders & Grevesse abundances
; are already included in the emissivities. In such cases, either
; leave out the atomic numbers (Z) in the call to LINEFLX() below,
; or redfine the abundance array to be relative to the APED values,
; e.g.,
if strpos(strlowcase(!LDBDIR),'aped',0) ge 0 then $
v_ABUND = !ABUND/getabund('anders & grevesse')
linint=lineflx(lconf,!LOGT,LWVL,Z,DEM=!DEM,abund=v_ABUND) ;[ph/s]
B] Read in continuum emissivities and calculate continuum intensities
; We can read in continuum emissivities using rd_cont().
; It is important to note that the output emissivities of rd_cont()
; are in [1e-23 erg cm^3/s/Ang] and not [1e-23 erg cm^3/s] as with rd_line()
; NOTE: To avoid multiple reads of the continuum emissivity database,
; we shall read in the emissivities over the entire range of interest
cconf=rd_cont(!CEROOT,n_e=!EDENS,$
wrange=[min(pspc_wvlar)<!WMIN,max(pspc_wvlar)>!WMAX],$
dbdir=!CDBDIR,abund=!ABUND,verbose=!VERBOSE,$
wvl=CWW,logT=ClogT)
; The continuum intensities per angstrom can be calculated again using
; lineflx(). Note that CWW contains the wavelength bin boundaries for
; the emissivity array.
CWVL=0.5*(CWW[1:*]+CWW)
conint=lineflx(cconf,!LOGT,CWVL,DEM=!DEM) ;[ph/s/Ang]
; Now to get just continuum intensity, we must multiply by an array
; containing the bin widths. If we define this array simply
; with: CDW=CWW[1:*]-CWW, we will get an ugly 'saw-toothed' figure.
; (a side-effect of the way the data-base is constructed) To work
; around this, we can use CWVL, the mid-bin values, and mid2bound(),
; which gives intelligent bin-boundary values given mid-bin values:
CWB=mid2bound(CWVL) & CDW=CWB[1:*]-CWB
conint=conint*CDW ;[ph/s/Ang]*[Ang]
C] Correct for inter-stellar absorption
; Derive ISM absorptions using ismtau()
ltau=ismtau(LWVL,NH=!NH,fH2=!fH2,He1=!He1,HeII=!HeII,$
Fano=Fano,wam=wam,/bam,abund=!ABUND,verbose=!VERBOSE)
ctau=ismtau(CWVL,NH=!NH,fH2=!fH2,He1=!He1,HeII=!HeII,$
Fano=Fano,wam=wam,/bam,abund=!ABUND,verbose=!VERBOSE)
ltrans=exp(-ltau) & ctrans=exp(-ctau)
; Derive theoretical line fluxes
linflx = linint * ltrans ;[ph/s/cm^2]
; Derive theoretical continuum fluxes
conflx = conint * ctrans ;[ph/s/cm^2]
D] Bin spectra and fold in effective area
; make input theoretical spectrum grid
nwbin_pspc = n_elements(pspc_effar)
dwvl=float((max(pspc_wvlar)-min(pspc_wvlar))/nwbin_pspc)
wgrid=findgen(nwbin+1L)*dwvl+min(pspc_wvlar)
; Rebin to form theoretical line spectrum using hastrogram()
linspc = hastogram(abs(LWVL),wgrid,wts=linflx) ;[ph/s/cm^2/bin]
; Rebin to form theoretical continuum spectrum using rebinw()
conspc = rebinw(conflx,CWVL,wgrid,/perbin) ;[ph/s/cm^2/bin]
; Derive predicted flux spectrum.
WVLS=0.5*(WGRID[1:*]+WGRID)
newEffAr=(interpol(pspc_effar,pspc_wvlar,WVLS) > 0) < (max(pspc_effar))
flxspc = (linspc + conspc) * newEffAr
; Derive predicted counts spectrum.
flxspc=flxspc*EXPTIME*1e3 ;[ct/bin]
; Restrict the counts spectrum to the specified ACIS-I range
oo = where(wgrid ge wmin_rosat and wgrid le wmax_rosat)
flxspc = flxspc(oo)
; Now get the total count rate and renormalize the DEM to the
; observed rate of 0.1 ct/s.
pred_rate = total(flxspc/EXPTIME/1e3) ;[ct/s]
print,''
if obs_rate gt 0 then $
print,'Rescaling input DEM by a factor '+strtrim(obs_rate/pred_rate,2)
print,''
rescale_factor=1.0
if obs_rate gt 0 then rescale_factor = obs_rate/pred_rate
!DEM = !DEM * rescale_factor
linint = linint * rescale_factor
conint = conint * rescale_factor
linflx = linflx * rescale_factor
conflx = conflx * rescale_factor
4. Construct XMM Spectra
; To construct the XMM spectra, we use the line and continuum
; emissivities read in above (LCONF and CCONF), the line and
; continuum intensities (LININT and CONINT) and fluxes (LINFLX
; and CONFLX) computed above, and recompute the predicted fluxes
; using XMM ARFs, and finally compute the observed spectra by
; convolving with the RMFs.
A] Read in line cooling emissivities and calculate line intensities
help,lconf,linint
B] Read in line cooling emissivities and calculate line intensities
help,cconf,conint
C] Correct for inter-stellar absorption
help,ltau,ltrans,linflx,ctau,ctrans,conflx
D] Bin spectra and fold in effective area
EMAX = !fundae.kevang/!WMIN & EMIN = !fundae.kevang/!WMAX
dnrg = float((EMAX-EMIN)/nwbin) ; bin size
egrid = findgen(nwbin+1L)*dnrg+EMIN ; bin boundaries [keV]
emid = 0.5*(egrid[1:*]+egrid) ; mid-bin values [keV]
wvls = !fundae.kevang/emid ; [ang]
linspc = hastogram(!fundae.kevang/abs(LWVL),egrid,wts=linflx) ; [ph/s/cm^2/bin]
conspc = rebinw(conflx,!fundae.kevang/CWB,egrid,/perbin) ; [ph/s/cm^2/bin]
;Read in the effective areas using rdarf()
if strlowcase(pn_ARF) eq 'none' then effar_pn=0*emid+1. else $
effar_pn = rdarf(pn_ARF,pn_ARF_str)
if strlowcase(mos_ARF) eq 'none' then effar_mos=0*emid+1. else $
effar_mos = rdarf(mos_ARF,mos_ARF_str)
if strlowcase(rgs1_ARF) eq 'none' then effar_rgs1=0*emid+1. else $
effar_rgs1 = rdarf(rgs1_ARF,rgs1_ARF_str)
if strlowcase(rgs2_ARF) eq 'none' then effar_rgs2=0*emid+1. else $
effar_rgs2 = rdarf(rgs2_ARF,rgs2_ARF_str)
;figure out the wavelength grid for effective areas
if n_tags(pn_ARF_str) eq 0 then nrgar_pn=emid else $
nrgar_pn = (0.5*(pn_ARF_str.ELO +pn_ARF_str.EHI))
if n_tags(mos_ARF_str) eq 0 then nrgar_mos=emid else $
nrgar_mos = (0.5*(mos_ARF_str.ELO+mos_ARF_str.EHI))
if n_tags(rgs1_ARF_str) eq 0 then nrgar_rgs1=emid else $
nrgar_rgs1 = (0.5*(rgs1_ARF_str.ELO+rgs1_ARF_str.EHI))
if n_tags(rgs2_ARF_str) eq 0 then nrgar_rgs2=emid else $
nrgar_rgs2 = (0.5*(rgs2_ARF_str.ELO+rgs2_ARF_str.EHI))
;interpolate to put effective area on binned spectra grids
new_effar_pn = (interpol(effar_pn ,nrgar_pn ,EMID) > 0) < (max(effar_pn))
new_effar_mos = (interpol(effar_mos ,nrgar_mos ,EMID) > 0) < (max(effar_mos))
new_effar_rgs1 = (interpol(effar_rgs1,nrgar_rgs1,EMID) > 0) < (max(effar_rgs1))
new_effar_rgs2 = (interpol(effar_rgs2,nrgar_rgs2,EMID) > 0) < (max(effar_rgs2))
;[ct/s/bin] (if DEM is [cm-5]: [ct/s/cm2/bin])
flxspc_pn = (linspc + conspc) * new_effar_pn
flxspc_mos = (linspc + conspc) * new_effar_mos
flxspc_rgs1 = (linspc + conspc) * new_effar_rgs1
flxspc_rgs2 = (linspc + conspc) * new_effar_rgs2
;Derive predicted counts spectrum
flxspc_pn = flxspc_pn *EXPTIME*1e3 ;[ct/bin]
flxspc_mos = flxspc_mos *EXPTIME*1e3 ;[ct/bin]
flxspc_rgs1 = flxspc_rgs1 *EXPTIME*1e3 ;[ct/bin]
flxspc_rgs2 = flxspc_rgs2 *EXPTIME*1e3 ;[ct/bin]
E] Convolve with RMF using
; read in RMFs
pn_RMF_str=rd_ogip_rmf(pn_RMF)
mos_RMF_str=rd_ogip_rmf(mos_RMF)
rgs1_RMF_str=rd_ogip_rmf(rgs1_RMF)
rgs2_RMF_str=rd_ogip_rmf(rgs2_RMF)
conv_rmf, egrid, flxspc_pn, CHAN_pn, CTSPC_pn, pn_RMF_str
conv_rmf, egrid, flxspc_mos, CHAN_mos, CTSPC_mos, mos_RMF_str
conv_rmf, egrid, flxspc_rgs1, CHAN_rgs1, CTSPC_rgs1, rgs1_RMF_str
conv_rmf, egrid, flxspc_rgs2, CHAN_rgs2, CTSPC_rgs2, rgs2_RMF_str
; Get co-added RGS spectrum.
CHAN_rgs = CHAN_rgs1 ;[keV]
CTSPC_rgs = CTSPC_rgs1 + CTSPC_rgs2 ;[ct/bin] valid grids same
flxspc_rgs = flxspc_rgs1+flx_spc_rgs2 ;[ct/bin] (if sperate ARF)
; Restrict the spectra to the specified range
oo_rgs = where(CHAN_rgs gt emin_xmm and CHAN_rgs lt emax_xmm)
oo_mos = where(CHAN_mos gt emin_xmm and CHAN_mos lt emax_xmm)
oo_pn = where(CHAN_pn gt emin_xmm and CHAN_pn lt emax_xmm)
CTSPC_rgs1 = CTSPC_rgs1(oo_rgs) & CHAN_rgs1 = CHAN_rgs1(oo_rgs)
CTSPC_rgs2 = CTSPC_rgs2(oo_rgs) & CHAN_rgs2 = CHAN_rgs2(oo_rgs)
CTSPC_rgs = CTSPC_rgs(oo_rgs) & CHAN_rgs = CHAN_rgs(oo_rgs)
CTSPC_mos = CTSPC_mos(oo_mos) & CHAN_mos = CHAN_mos(oo_mos)
CTSPC_pn = CTSPC_mos(oo_pn) & CHAN_pn = CHAN_pn(oo_pn)
; Note that the output energy grid of the spectra will be by
; default the energy grid defined by the RMF. The spectrum
; however will only show lines and continuum between the
; selected wavelength ranges.
5. Obtain Poisson deviates and plot
; The final step is a simulation of counts based on the spectrum
; predicted above
nbin_pn = n_elements(CTSPC_pn) & CTSIM_pn = intarr(nbin_pn)
nbin_mos = n_elements(CTSPC_mos) & CTSIM_mos = intarr(nbin_mos)
nbin_rgs = n_elements(CTSPC_rgs) & CTSIM_rgs = intarr(nbin_rgs)
for i=0L,nbin_pn-1L do if CTSPC_pn[i] gt 0 then $
CTSIM_pn[i] =randomu(seed,poisson=CTSPC_pn[i])
for i=0L,nbin_mos-1L do if CTSPC_mos[i] gt 0 then $
CTSIM_mos[i]=randomu(seed,poisson=CTSPC_mos[i])
for i=0L,nbin_rgs-1L do if CTSPC_rgs[i] gt 0 then $
CTSIM_rgs[i]=randomu(seed,poisson=CTSPC_rgs[i])
; The results of the calculations are plotted below
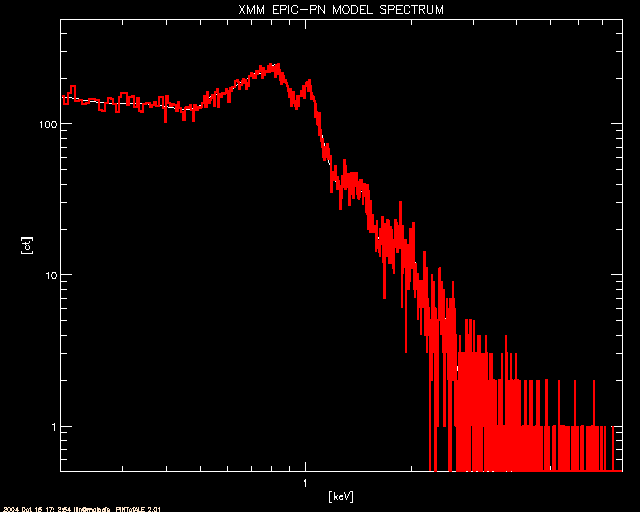
; The EPIC-pn model (in white) and simulated counts (in red)
window, 0
plot, CHAN_pn, CTSPC_pn, title='XMM EPIC-PN MODEL SPECTRUM',/xl,/yl,$
xtitle='[keV]', ytitle='[ct]', ystyle=1, xstyle=1, $
xrange=[emin_xmm,emax_xmm], yrange=[0.5,2*max(CTSPC_pn)] & stample
oplot, CHAN_pn, CTSIM_pn ,color = 2, psym=10, thick=2
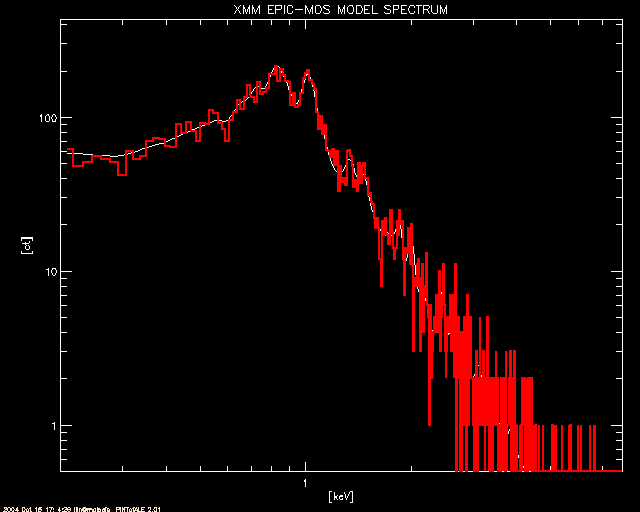
 ; The EPIC-mos model (in white) and simulated counts (in red)
window, 1
plot, CHAN_mos, CTSPC_mos, title='XMM EPIC-MOS MODEL SPECTRUM',/xl,/yl,$
xtitle='[keV]', ytitle='[ct]', ystyle=1, xstyle=1, $
xrange=[emin_xmm,emax_xmm], yrange=[0.5,2*max(CTSPC_mos)] & stample
oplot, CHAN_mos, CTSIM_mos, color = 2, psym=10, thick=2
; The EPIC-mos model (in white) and simulated counts (in red)
window, 1
plot, CHAN_mos, CTSPC_mos, title='XMM EPIC-MOS MODEL SPECTRUM',/xl,/yl,$
xtitle='[keV]', ytitle='[ct]', ystyle=1, xstyle=1, $
xrange=[emin_xmm,emax_xmm], yrange=[0.5,2*max(CTSPC_mos)] & stample
oplot, CHAN_mos, CTSIM_mos, color = 2, psym=10, thick=2
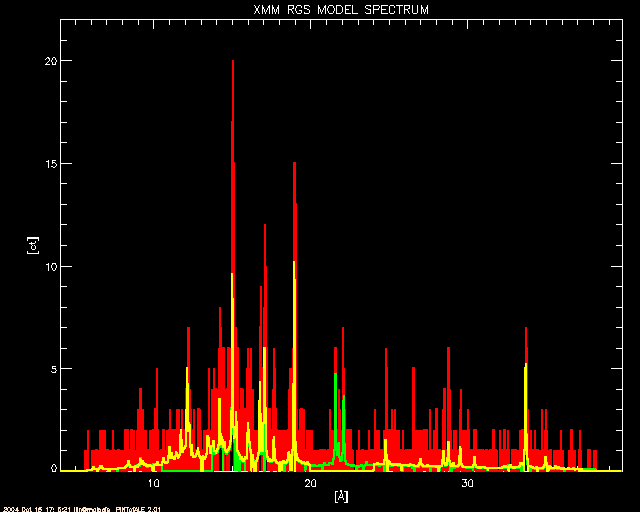
 ; The RGS model (in white) and simulated counts (in red). Also shown
; are the model counts from RGS1 (yellow) and RGS2 (green)
window, 2
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,1.1*max(CTSIM_rgs)]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10, thick=2
; The RGS model (in white) and simulated counts (in red). Also shown
; are the model counts from RGS1 (yellow) and RGS2 (green)
window, 2
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,1.1*max(CTSIM_rgs)]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10, thick=2
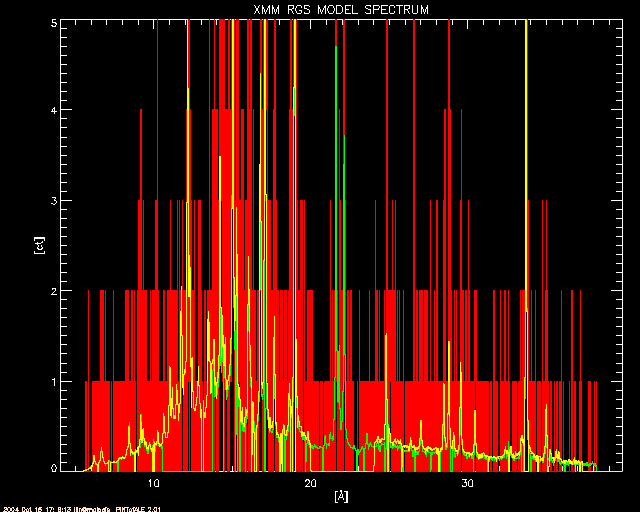
 ; As above, but zoomed in to show the low counts detail.
window, 3
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,5.*median(CTSPC_rgs)>5.]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10
; As above, but zoomed in to show the low counts detail.
window, 3
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,5.*median(CTSPC_rgs)>5.]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10
 ; We may summarize results as follows:
;
; Simulated counts spectra are in:
; CTSIM_pn(chan_pn),CTSIM_mos(CHAN_mos),CTSIM_rgs(CHAN_rgs) [ct/bin]
;
; Redistributed counts spectra are in:
; CTSPC_pn(chan_pn),CTSPC_mos(CHAN_mos),CTSPC_rgs(CHAN_rgs) [ct/bin]
;
; Predicted counts spectra are in:
; FLXSPC_pn(wvls),FLXSPC_mos(wvls), FLXSPC_rgs(wvls) [ct/bin]
;
; Note: Standard RGS response matrices include the effective area, so, unless
; a seperate rmf and effective area files are used, FLXSPC_rgs will
; NOT contain predicted counts spectra.
;
; Theoretical line fluxes are in:
; linspc(lwvl) [ph/s[/cm^2]]
;
; Theoretical continuum fluxes are in:
; conspc(cwvl) [ph/s[/cm^2]]
; We may summarize results as follows:
;
; Simulated counts spectra are in:
; CTSIM_pn(chan_pn),CTSIM_mos(CHAN_mos),CTSIM_rgs(CHAN_rgs) [ct/bin]
;
; Redistributed counts spectra are in:
; CTSPC_pn(chan_pn),CTSPC_mos(CHAN_mos),CTSPC_rgs(CHAN_rgs) [ct/bin]
;
; Predicted counts spectra are in:
; FLXSPC_pn(wvls),FLXSPC_mos(wvls), FLXSPC_rgs(wvls) [ct/bin]
;
; Note: Standard RGS response matrices include the effective area, so, unless
; a seperate rmf and effective area files are used, FLXSPC_rgs will
; NOT contain predicted counts spectra.
;
; Theoretical line fluxes are in:
; linspc(lwvl) [ph/s[/cm^2]]
;
; Theoretical continuum fluxes are in:
; conspc(cwvl) [ph/s[/cm^2]]
6. Comparison with PIMMS
; Results using this thread are roughly consistent with
; PIMMS results. PIMMS returned the following XMM count rates
; for a ROSAT/PSPC count rate of 0.1 cts/s and a Raymond-Smith
; model of temperature 0.5437 keV:
; XMM PN MED : 7.792E-01 cts/s
; XMM MOS MED : 1.958E-01 cts/s
; XMM RGS1 O1 : 2.629E-02 cts/s
; XMM RGS2 O1 : 3.627E-02 cts/s
;
; Running this thread with !ABUND = getabund('Allen'),
; T_components = [6.8], and obs_rate = 0.1 gives:
; XMM PN MED : 0.74 cts/s
; XMM MOS MED : 0.19 cts/s
; XMM RGS1 O1 : 0.027 cts/s
; XMM RGS2 O1 : 0.035 cts/s
pintofale()head.cfa.harvard.edu (LL)
 ; The EPIC-mos model (in white) and simulated counts (in red)
window, 1
plot, CHAN_mos, CTSPC_mos, title='XMM EPIC-MOS MODEL SPECTRUM',/xl,/yl,$
xtitle='[keV]', ytitle='[ct]', ystyle=1, xstyle=1, $
xrange=[emin_xmm,emax_xmm], yrange=[0.5,2*max(CTSPC_mos)] & stample
oplot, CHAN_mos, CTSIM_mos, color = 2, psym=10, thick=2
; The EPIC-mos model (in white) and simulated counts (in red)
window, 1
plot, CHAN_mos, CTSPC_mos, title='XMM EPIC-MOS MODEL SPECTRUM',/xl,/yl,$
xtitle='[keV]', ytitle='[ct]', ystyle=1, xstyle=1, $
xrange=[emin_xmm,emax_xmm], yrange=[0.5,2*max(CTSPC_mos)] & stample
oplot, CHAN_mos, CTSIM_mos, color = 2, psym=10, thick=2
 ; The RGS model (in white) and simulated counts (in red). Also shown
; are the model counts from RGS1 (yellow) and RGS2 (green)
window, 2
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,1.1*max(CTSIM_rgs)]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10, thick=2
; The RGS model (in white) and simulated counts (in red). Also shown
; are the model counts from RGS1 (yellow) and RGS2 (green)
window, 2
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,1.1*max(CTSIM_rgs)]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10, thick=2
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10, thick=2
 ; As above, but zoomed in to show the low counts detail.
window, 3
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,5.*median(CTSPC_rgs)>5.]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10
; As above, but zoomed in to show the low counts detail.
window, 3
plot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, title='XMM RGS MODEL SPECTRUM',$
xtitle='['+!AA+']', ytitle='[ct]', ystyle=1, xstyle=1, /nodata,$
xrange=[!fundae.kevang/MAX(CHAN_rgs),!fundae.kevang/MIN(CHAN_rgs)],$
yrange=[0,5.*median(CTSPC_rgs)>5.]
oplot, !fundae.kevang/CHAN_rgs, CTSIM_rgs, color=2, psym=10
oplot, !fundae.kevang/CHAN_rgs1, CTSPC_rgs1, color=3, psym=10
oplot, !fundae.kevang/CHAN_rgs2, CTSPC_rgs2, color=4, psym=10
 ; We may summarize results as follows:
;
; Simulated counts spectra are in:
; CTSIM_pn(chan_pn),CTSIM_mos(CHAN_mos),CTSIM_rgs(CHAN_rgs) [ct/bin]
;
; Redistributed counts spectra are in:
; CTSPC_pn(chan_pn),CTSPC_mos(CHAN_mos),CTSPC_rgs(CHAN_rgs) [ct/bin]
;
; Predicted counts spectra are in:
; FLXSPC_pn(wvls),FLXSPC_mos(wvls), FLXSPC_rgs(wvls) [ct/bin]
;
; Note: Standard RGS response matrices include the effective area, so, unless
; a seperate rmf and effective area files are used, FLXSPC_rgs will
; NOT contain predicted counts spectra.
;
; Theoretical line fluxes are in:
; linspc(lwvl) [ph/s[/cm^2]]
;
; Theoretical continuum fluxes are in:
; conspc(cwvl) [ph/s[/cm^2]]
; We may summarize results as follows:
;
; Simulated counts spectra are in:
; CTSIM_pn(chan_pn),CTSIM_mos(CHAN_mos),CTSIM_rgs(CHAN_rgs) [ct/bin]
;
; Redistributed counts spectra are in:
; CTSPC_pn(chan_pn),CTSPC_mos(CHAN_mos),CTSPC_rgs(CHAN_rgs) [ct/bin]
;
; Predicted counts spectra are in:
; FLXSPC_pn(wvls),FLXSPC_mos(wvls), FLXSPC_rgs(wvls) [ct/bin]
;
; Note: Standard RGS response matrices include the effective area, so, unless
; a seperate rmf and effective area files are used, FLXSPC_rgs will
; NOT contain predicted counts spectra.
;
; Theoretical line fluxes are in:
; linspc(lwvl) [ph/s[/cm^2]]
;
; Theoretical continuum fluxes are in:
; conspc(cwvl) [ph/s[/cm^2]]